After the symposium yesterday, and several more insightful comments, I thought I should write a couple of quick points.
One of the main issues is penetrance, or how often the disease occurs when you have a given genomic profile. For some diseases, like Huntington's disease, having the particular mutation translates directly into a certainty that you will have the disease. There really isn't much of a chance that you'll somehow avoid developing it. For other diseases, a gene may change your likelihood of developing the disease slightly or in an almost un-noticeable way. In fact, sometimes you may have offsetting changes that negate what would be a risk factor in another person. Genomes are wild and complex data structures, and are definitely not digital in the sense that seeing a particular variation will always give you a certain result.
Mainly, that has to do with the biology of the cell. There are often redundant pathways to accomplish a given task, or several levels of regulation that can be called on to turn genes on or off. Off the top of my head, I can think of several levels of regulation (dna methylation, histone post-translational modifications, enhancers, promotors, microrna, ubiquitination leading to increased degradation, splicing, mis-folding through chaperonin regulation, etc) that can be used to fine tune or throttle the systems in any given cell. At that rate, looking at a single variation seems like it might be an entirely useless venture.
And, in fact, that was the general consensus of the panelists last night: the companies that currently run a microarray on your dna and then report to you some slight changes in risk factors are really a waste of time - they don't begin to compensate for the complexity that is really going on.
However, my contention isn't that we should be doing personal medicine over the whole genome, but that as we move forward, that personal medicine will have a large and growing impact over how healthcare is practiced. I've heard several people talk about Warfarin as an example of this. Warfarin is used to treat hypertension, and is quite effective in most people. However, each person has different dosage requirements - not because they need more to activate the pathway, but because we all degrade it at different rates, depending on which p450 enzymes we have to break it down.
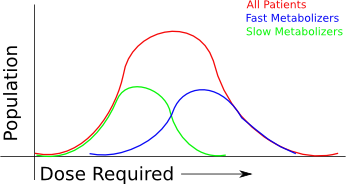
In the above graph, you can see all patients conform to some "normal" distribution, but they're really made up of two subpopulations - one set of fast metabolizers and one set of slow metabolizers, as judged by metabolism of some other drugs. (Yes, I'm way oversimplifying how this works - this is not real warfarin data!) When you look at the spectrum of patients that come in, you see a continuum of patient dosages, but you'd never understand why.
Instead, you could look for markers. In the case of drug metabolism, only one p450 may be responsible for the speed at which the drug is processed, so looking at the same group of patients for that particular trait will give you a completely different graph:

Which means, you can start to figure out what initial dose will be required, and tweak from there.
(If you're wondering why the fast metabolizers and slow metabolizers of the same drug have some overlap in my example, it's just so I'd have an excuse to say there are probably other factors involved: environment, other things interfering with the metabolism, the rate at which the kidneys clear the drugs... and probably many other things I've never considered.)
So what's my point? It's easy. Personal medicine isn't about whole genomics, but rather about finding out what conditions underly the complex behaviours of the body - and then to apply that knowledge as best as we can to treat people. (Whole Genome Studies will be important to learning how these things work though, so without the ability to do whole genome sequencing, we wouldn't have a chance at really making personal medicine effective.) I'll be the first to admit we don't know enough to do this for all diseases, but we certainly do know enough to begin applying it to a few. I've argued that within 5 years, we'll start to really see the effects. It won't be a radical change to all medical care at once, but a slow progression into the clinics.
To narrow my prediction down further, at some point in the next 5 years, it will become routine (~10-20% of patients?) for doctors to start doing genomic tests (not full genome sequencing!) to apply this type of knowledge when they treat their patients with new drugs. (Not every illness will require genomic information, so clearly we'll never reach 100% requirement for it - having a splinter removed in the E.R. won't require the doc to check your genome...) I give it another 10 years before full genome sequencing begins hitting clinics.. and even that will be a gradual change.
Now I've really wandered far outside of my field. I'll let the doctors and physicians handle it from here and try to restrict my comments to the more scientific aspects of it.
Labels: biochemistry, personalized medicine



